Papers, or it didn't happen.
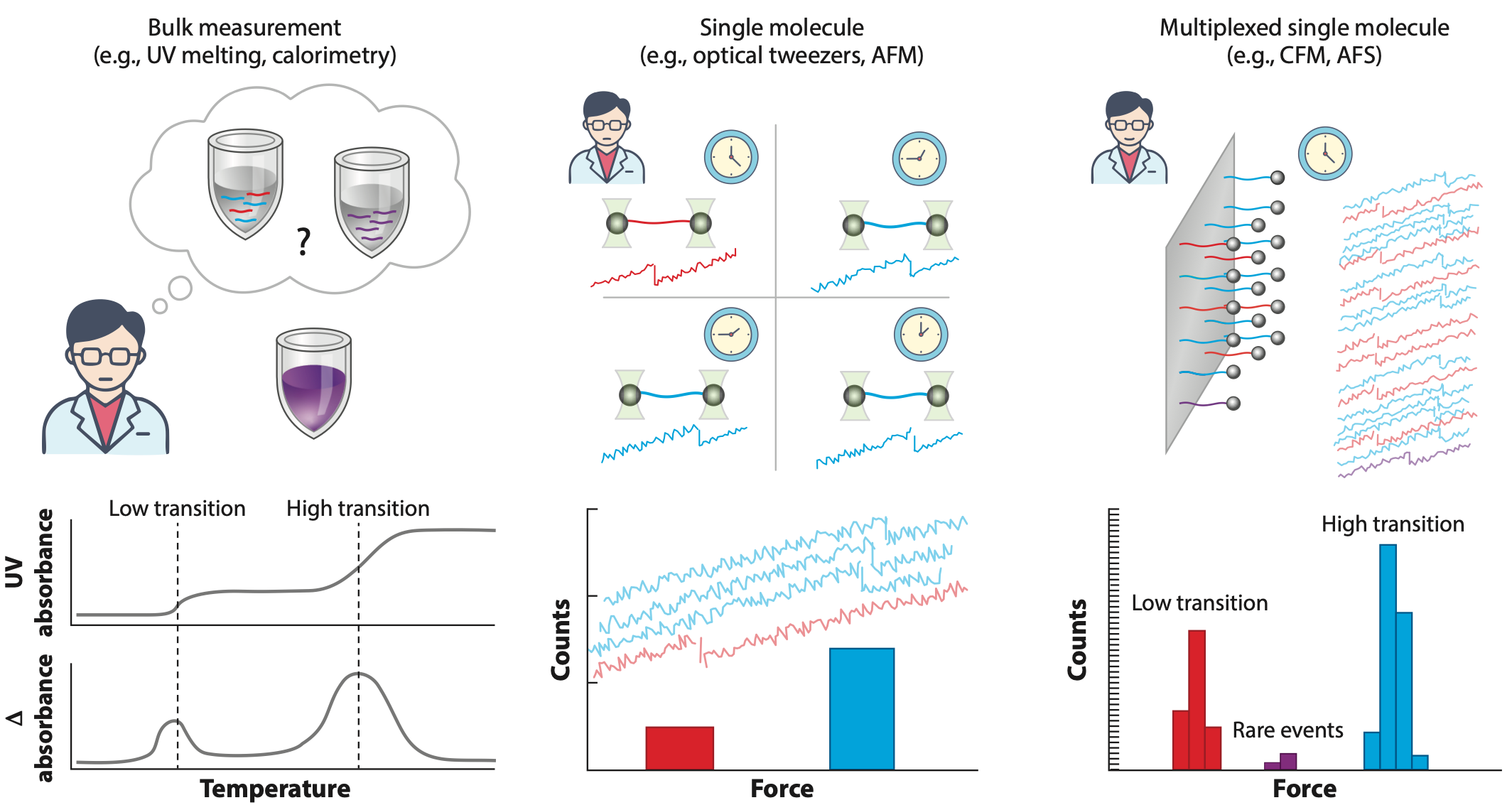
Beyond the single molecule: Multiplexed methods in force spectroscopy
K Halvorsen, A Ward, WP Wong. Annual Review of Biophysics (2026). [PDF]
Single-molecule techniques have transformed biological research by en- abling direct observation and manipulation of individual molecules. These methods overcome ensemble averaging inherent in bulk measurements and facilitate studies under physiological stresses and out-of-equilibrium conditions. They have provided valuable insights into diverse biological processes, from stepping mechanisms of molecular motors to mechanical properties of biomolecules to the dynamic strength of intermolecular bonds. Advances in multiplexed and high-throughput single-molecule force spectroscopy methods are improving throughput, capabilities, and accessibility. In this review, we detail the evolution of multiplexed force spectroscopy technologies, highlighting key advances in instrumentation, molecular engineering, and analytical techniques. We discuss diverse applications spanning molecular biophysics, biomolecular sensing, proteomics, and cellular mechanobiology. Finally, we explore ongoing challenges and future opportunities and highlight how the impact of multiplexed single-molecule force spectroscopy can continue to grow through further developments in novel instrumentation, chemical tools, and innovative applications.
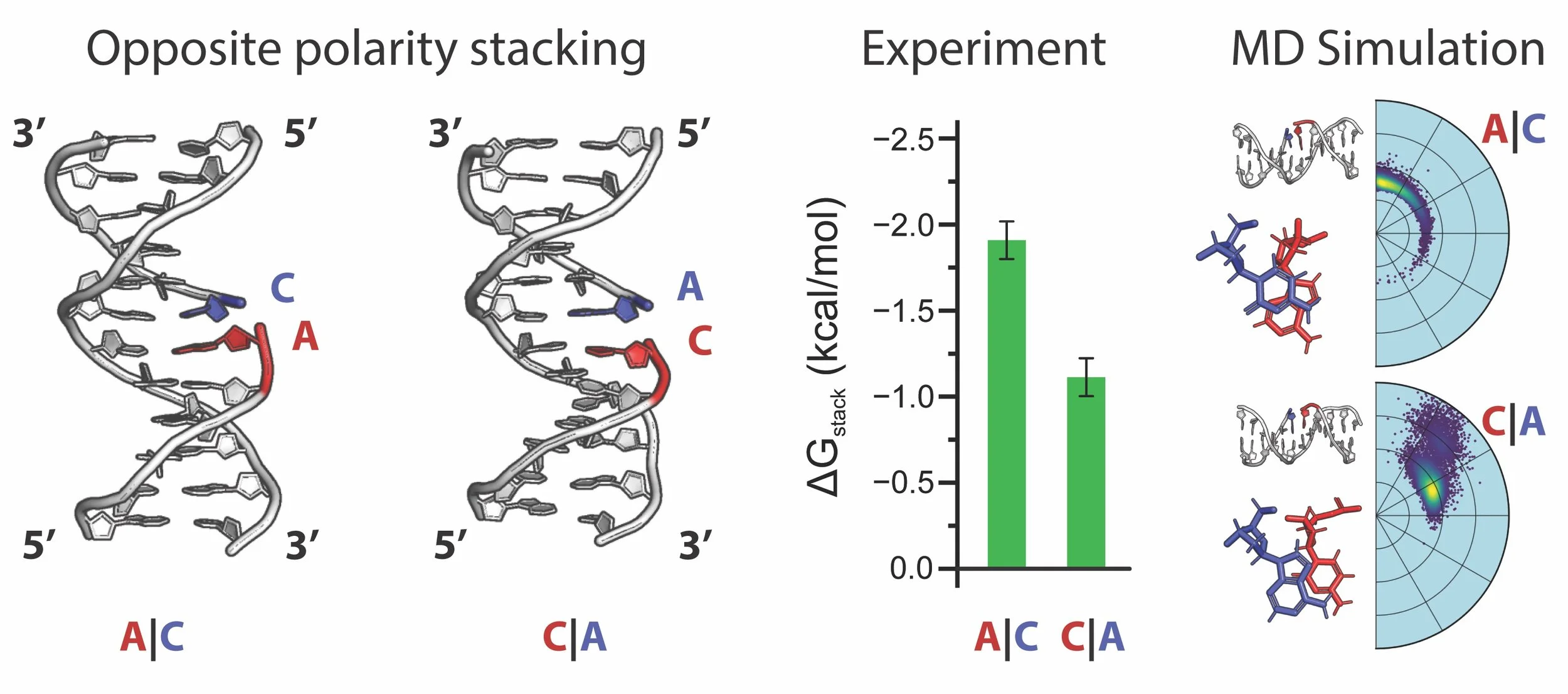
Investigating polarity effects in DNA base stacking
JA Punnoose, CS Kam, T Melfi, S Vengaveti, AA Chen, K Halvorsen. JACS Au (2025). [PDF]
Nucleic acid structures are stabilized by both base pairing and base stacking. While energetics of base pairing interactions are relatively well established, our understanding of the energetic contributions of base stacking remain incomplete. Here, we use a combination of single-molecule and computational biophysics approaches to investigate the effect of strand polarity on base-stacking energetics. We designed pairs of DNA constructs with reversed stacking polarities at nick sites, along with corresponding no-stack controls to isolate stacking contributions. Performing single-molecule force-clamp assays with a Centrifuge Force Microscope (CFM), we observed polarity-dependent differences in stacking energetics. These differences were most pronounced in purine–purine and certain purine–pyrimidine interactions. Notably, a 5′ purine stacked on a 3′ pyrimidine was generally more stable than the reverse polarity. We employed molecular dynamics (MD) simulations to observe stacking interfaces in the DNA constructs. The simulations were qualitatively consistent with our experiments, and showed positional differences between opposite polarity stacking pairs, giving some insight into the origin of these polarity differences. Overall, these results demonstrate that base polarity can modulate stacking stability and should be considered when designing short duplex regions such as overhangs in molecular biology and biotechnology applications.
Counterions influence the isothermal self-assembly of DNA nanostructures
A Rodriguez, BR Madhanagopal, K Sarkar, Z Nowzari, J Mathivanan, H Talbot, A Patel, V Morya, K Halvorsen, S Vangaveti, JA Berglund, AR Chandrasekaran. Science Advances (2025). [PDF]
DNA nanostructures are typically assembled by thermal annealing in buffers containing magnesium. We demonstrate the assembly of DNA nanostructures at constant temperatures ranging from 4° to 50°C in solutions containing different counterions. The choice of counterions and the assembly temperature influence the isothermal assembly of several DNA motifs and designed three-dimensional DNA crystals. Molecular dynamics simulations show more fluctuations of the DNA structure in select monovalent ions (Na+ and K+) compared to divalent ions (Mg2+and Ca2+). A key highlight is the successful assembly of DNA motifs in nickel-containing buffer at temperatures below 40°C, otherwise unachievable at higher temperatures or using an annealing protocol. DNA nanostructures isothermally assembled in different ions do not affect the viability of fibroblasts, myoblasts, and myotubes or the immune response in myoblasts. The use of ions other than the typically used magnesium holds key potential in biological and materials science applications that require minimal amounts of magnesium.
Tuning the stability of DNA tetrahedra with base stacking interactions
JA Punnoose, D Cole, T Melfi, V Morya, BR Madhanagopal, AA Chen, S Vangaveti, AR Chandrasekaran, K Halvorsen. Nano Letters (2025). [PDF]
DNA nanotechnology uses the programmable assembly of DNA to create nanoscale objects. Recent work from our laboratory suggested that terminal stacking interactions between adjacent strands could be a design parameter for DNA nanotechnology. Here, we explore that idea by creating DNA tetrahedra with sticky ends containing identical base pairing interactions but different stacking interactions. Testing all 16 stacking combinations, we found that the melting temperature of DNA tetrahedra varied by up to 10 °C from altering a single base stack in the design. We also show that a 4 bp sticky end with weak stacking does not form stable tetrahedra, while strengthening the stacks confers high stability with a 46.8 ± 1.2 °C melting temperature, comparable to that of a 6 bp sticky end with weak stacking (49.7 ± 2.9 °C). The results likely apply to other DNA nanostructures and suggest that stacking interactions play a role in the formation and stability of DNA nanostructures.
A guide to building a low-cost centrifuge force microscope module for single-molecule force experiments
JA Punnoose, A Hayden, CS Kam, K Halvorsen. Nature Protocols (2024). [PDF]
The ability to apply controlled forces to individual molecules or molecular complexes and observe their behaviors has led to many important discoveries in biology. Instruments capable of probing single-molecule forces typically cost >US$100,000, limiting the use of these techniques. The centrifuge force microscope (CFM) is a low-cost and easy-to-use instrument that enables high- throughput single-molecule studies. By combining the imaging capabilities of a microscope with the force application of a centrifuge, the CFM enables the simultaneous probing of hundreds to thousands of single-molecule interactions using tethered particles. Here we present a comprehensive set of instructions for building a CFM module that fits within a commercial benchtop centrifuge. The CFM module uses a 3D-printed housing, relies on off-the-shelf optical and electrical components, and can be built for less than US$1,000 in about 1 day. We also provide detailed instructions for setting up and running an experiment to measure force-dependent shearing of a short DNA duplex, as well as the software for CFM control and data analysis. The protocol is suitable for users with basic experience in analytical biochemistry and biophysics. The protocol enables the use of CFM-based experiments and may facilitate access to the single-molecule research field.
Resolving altered base-pairing of RNA modifications with DNA nanoswitches
IA Todkari, AR Chandrasekaran, JA Punnoose, S Mao, P Haruehanroengra, C Beckles, J Sheng, K Halvorsen. Nucleic Acids Research (2023). [PDF]
There are >170 naturally occurring RNA chemical modifications, with both known and unknown biological functions. Analytical methods for detecting chemical modifications and for analyzing their effects are relatively limited and have had difficulty keeping pace with the demand for RNA chemical biology and biochemistry research. Some modifications can affect the ability of RNA to hybridize with its complementary sequence or change the selectivity of base pairing. Here, we investigate the use of affinity-based DNA nanoswitches to resolve energetic differences in hybridization. We found that a single m3C modification can sufficiently destabilize hybridization to abolish a detection signal, while an s4U modification can selectively hybridize with G over A. These results establish proof of concept for using DNA nanoswitches to detect certain RNA modifications and analyzing their effects in base pairing stability and specificity.
Fluorometric determination of DNA nanostructure biostability
H Talbot, BR Madhanagopal, A Hayden, K Halvorsen, AR Chandrasekaran. ACS Applied Bio Materials (2023). [PDF]
The analysis and improvement of DNA nanostructure biostability is one of the keys areas of progress needed in DNA nanotechnology applications. Here, we present a plate-compatible fluorometric assay for measuring DNA nanostructure biostability using the common intercalator ethidium bromide. We demonstrate the assay by testing the biostability of duplex DNA, a double crossover DNA motif, and a DNA origami nanostructure against different nucleases and in fetal bovine serum. This method scales well to measure a large number of samples using a plate reader and can complement existing methods for assessing and developing robust DNA nanostructures.
The role of size in biostability of DNA tetrahedra
J Vilcapoma, A Patel, AR Chandrasekaran, K Halvorsen. Chemical Communications (2023). [PDF]
The potential for using DNA nanostructures for drug delivery applications requires understanding and ideally tuning their biostability. Here we investigate how biological degradation varies with size of a DNA nanostructure. We designed DNA tetrahedra of three edge lengths ranging from 13 to 20 bp and analyzed nuclease resistance for two nucleases and biostability in fetal bovine serum. We found that DNase I had similar digestion rates across sizes but appeared to incompletely digest the smallest tetrahedron, while T5 exonuclease was notably slower to digest the largest tetrahedron. In fetal bovine serum, the 20 bp tetrahedron was degraded four times faster than the 13 bp. These results show that DNA nanostructure size can influence nuclease degradation, but suggest a complex relationship that is nuclease specific.
High-throughput single-molecule quantification of individual base stacking energies in nucleic acids
JA Punnoose, KJ Thomas, AR Chandrasekaran, J Vilcapoma, A Hayden, K Kilpatrick, S Vangaveti, A Chen, T Banco, K Halvorsen. Nature Communications 14, 631 (2023). [PDF]
Base stacking interactions between adjacent bases in DNA and RNA are important for many biological processes and in biotechnology applications. Previous work has estimated stacking energies between pairs of bases, but contributions of individual bases has remained unknown. Here, we use a Centrifuge Force Microscope for high-throughput single molecule experiments to measure stacking energies between adjacent bases. We found stacking energies strongest between purines (G|A at −2.3 ± 0.2 kcal/mol) and weakest between pyrimidines (C|T at −0.5 ± 0.1 kcal/mol). Hybrid stacking with phosphorylated, methylated, and RNA nucleotides had no measurable effect, but a fluorophore modification reduced stacking energy. We experimentally show that base stacking can influence stability of a DNA nanostructure, modulate kinetics of enzymatic ligation, and assess accuracy of force fields in molecular dynamics simulations. Our results provide insights into fundamental DNA interactions that are critical in biology and can inform design in biotechnology applications.
Encoding, decoding, and rendering information in DNA nanoswitch libraries
H Talbot, K Halvorsen, AR Chandrasekaran. ACS Synthetic Biology (2022). [PDF]
DNA-based construction allows the creation of molecular devices that are useful in information storage and processing. Here, we combine the programmability of DNA nanoswitches and stimuli-responsive conformational changes to demonstrate information encoding and graphical readout using gel electrophoresis. We encoded information as 5-bit binary codes for alphanumeric characters using a combination of DNA and RNA inputs that can be decoded using molecular stimuli such as a ribonuclease. We also show that a similar strategy can be used for graphical visual readout of alphabets on an agarose gel, information that is encoded by nucleic acids and decoded by a ribonuclease. Our method of information encoding and processing could be combined with DNA actuation for molecular computation and diagnostics that require a nonarbitrary visual readout.
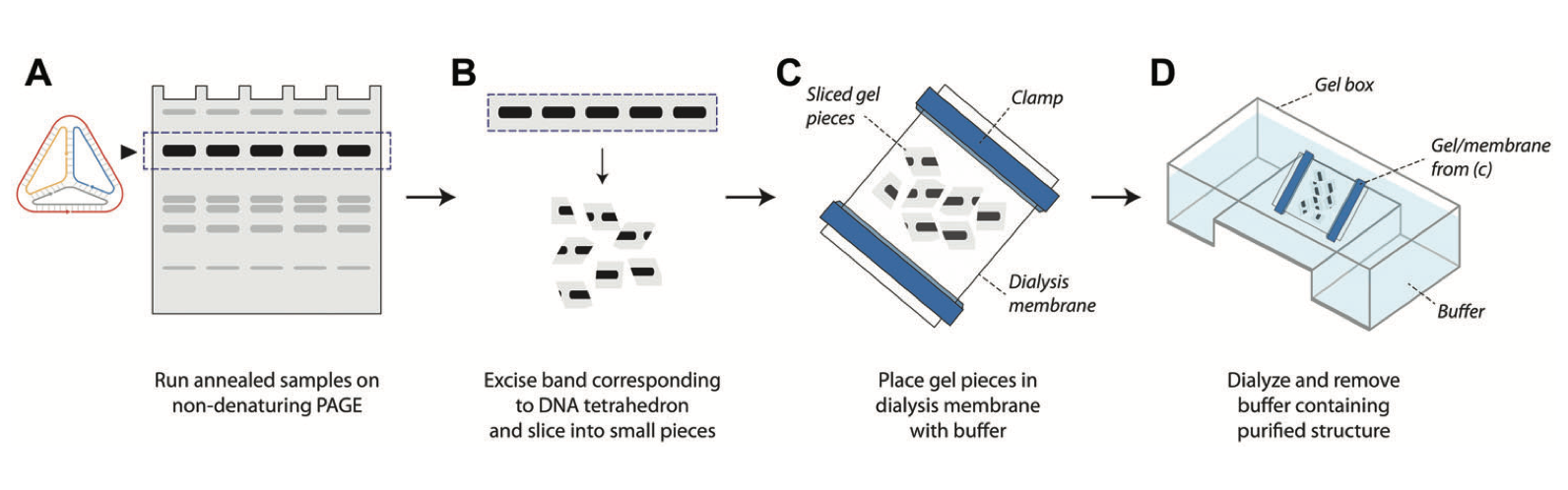
Purification of self-assembled DNA tetrahedra using gel electrophoresis
A Patel, VA Valsangkar, K Halvorsen, AR Chandrasekaran. Current Protocols in Nucleic Acid Chemistry, 2, e560 (2022). [PDF]
DNA nanostructures have found applications in a variety of fields such as biosensing, drug delivery, cellular imaging, and computation. Several of these applications require purification of the DNA nanostructures once they are assembled. Gel electrophoresis–based purification of DNA nanostructures is one of the methods used for this purpose. Here, we describe a step-by-step protocol for a gel-based method to purify self-assembled DNA tetrahedra. With further optimization, this method could also be adapted for other DNA nanostructures.
Surface-enhanced Raman spectroscopy for drug discovery: peptide-RNA binding
LM Almehmadi, VA Valsangkar, K Halvorsen, Q Zhang, J Sheng, IK Lednev. Analytical and Bioanalytical Chemistry, 414, 6009-6016 (2022). [PDF]
The ever-growing demand for new drugs highlights the need to develop novel cost- and time-effective techniques for drug discovery. Surface-enhanced Raman spectroscopy (SERS) is an emerging ultrasensitive and label-free technique that allows for the efficient detection and characterization of molecular interactions. We have recently developed a SERS platform for detecting a single protein molecule linked to a gold substrate (Almehmadi et al. Scientific Reports 2019). In this study, we extended the approach to probe the binding of potential drugs to RNA targets. To demonstrate the proof of concept, two 16-amino acid residue peptides with close primary structures and different binding affinities to the RNA CUG repeat related to myotonic dystrophy were tested. Three-microliter solutions of the RNA repeat with these peptides at nanomolar concentrations were probed using the developed approach, and the binding of only one peptide was demonstrated. The SER spectra exhibited significant fluctuations along with a sudden strong enhancement as spectra were collected consecutively from individual spots. Principal component analysis (PCA) of the SER spectral datasets indicated that free RNA repeats could be differentiated from those complexed with a peptide with 100% accuracy. The developed SERS platform provides a novel opportunity for label-free screening of RNA-binding peptides for drug discovery.
Single species RNA purification using DNA nanoswitches
AR Chandrasekaran, L Zhou, K Halvorsen. Trends Biochem. Sci. 47, 367-368 (2022). [PDF]
RNA purification is a critical need in biomedical research. The development of tools for analyzing structure, function, and chemical modifications of RNA have outpaced those for RNA purification. The DNA nanoswitch method is a new ‘catch and release’ approach for targeted purification of specific RNA sequences from complex mixtures. Capture of a target RNA reconfigures the DNA nanoswitch from the linear ‘off’ state to the looped ‘on’ state. The RNA bound to the nanoswitch can be quantified and isolated to obtain pure RNA of a single sequence. The method is useful in purifying specific microRNAs, ribosomal RNAs, and mRNAs from cellular total RNA extracts.
Sequence-selective purification of biological RNAs using DNA nanoswitches
L Zhou, A Hayden, AR Chandrasekaran, J Vilcapoma, C Cavaliere, P Dey, S Mao, J Sheng, BK Dey, P Rangan, K Halvorsen. Cell Reports Methods, 1, 100126 (2021). [PDF]
Nucleic acid purification is a critical aspect of biomedical research and a multibillion-dollar industry. Here we establish sequence-selective RNA capture, release, and isolation using conformationally responsive DNA nanoswitches. We validate purification of specific RNAs ranging in size from 22 to 401 nt with up to 75% recovery and 99.98% purity in a benchtop process with minimal expense and equipment. Our method compared favorably with bead-based extraction of an endogenous microRNA from cellular total RNA, and can be programmed for multiplexed purification of multiple individual RNA targets from one sample. Coupling our approach with downstream LC/MS, we analyzed RNA modifications in 5.8S ribosomal RNA, and found 2’-O-methylguanosine, 2’-O-methyluridine, and pseudouridine in a ratio of ~1:7:22. The simplicity, low cost, and low sample requirements of our method make it suitable for easy adoption, and the versatility of the approach provides opportunities to expand the strategy to other biomolecules.
DNA-Based Smart Reagent for Detecting Alzheimer’s Associated MicroRNAs
AR Chandrasekaran, K Halvorsen. ACS Sensors 446618 (2021). [PDF]
Alzheimer’s disease (AD) is the most common neurodegenerative disorder, with significant research efforts devoted to identifying new biomarkers for clinical diagnosis and treatment. MicroRNAs have emerged as likely disease regulators and biomarkers for AD, now implicated as having roles in several biological processes related to progression of the disease. In this work, we use the miRacles assay (microRNA activated conditional looping of engineered switches) for single-step detection of AD-related microRNAs. The technology is based on conformationally responsive DNA nanoswitches that loop upon recognition of a target microRNA and report their on/off status through an electrophoretic readout. Unlike many other methods, our approach directly detects native microRNAs without amplification or labeling, eliminating the need for expensive enzymes, reagents, and equipment. We used this assay to screen for AD-related microRNAs, demonstrate specificity within a microRNA family, sensitivity of ∼ 8 fM, and multiplexing capability to simultaneously detect four microRNA targets. Toward clinical use, we provide proof-of-concept detection and quantifiable dysregulation of specific microRNAs from total RNA extracts derived from healthy and AD brain samples. In the context of AD, this “smart reagent” could facilitate biomarker discovery, accelerate efforts to understand the role of microRNAs in AD, and have clinical potential as a diagnostic or monitoring tool for validated biomarkers.
A mini DNA-RNA hybrid origami nanobrick
L Zhou, AR Chandrasekaran, M Yan, VA Valsangkar, JI Feldblyum, J Sheng, K Halvorsen. Nanoscale Adv. DOI: 10.1039/D1NA00026H (2021). [PDF]
DNA origami is typically used to fold a long single-stranded DNA scaffold into nanostructures with complex geometries using many short DNA staple strands. Integration of RNA into nucleic acid nanostructures is also possible, but has been less studied. In this research, we designed and characterized a hybrid RNA-scaffolded origami nanostructure with dimensions of ~12 nm. We used 12 DNA staple strands to fold a 401 nt RNA scaffold into a ten-helix bundle with a honeycomb cross section. We verified the construction of the nanostructure using gel electrophoresis and atomic force microscopy. The DNA-RNA hybrid origami showed higher resistance to ribonuclease compared to a DNA-RNA duplex control. Our work shows potential use in folding long RNA, such as messenger RNA, into origami nanostructures that can be delivered into targeted cells as medicine or vaccine.
Orthogonal Control of DNA Nanoswitches with Mixed Physical and Biochemical Cues
NT Forrest, J Vilcapoma, K Alejos, K Halvorsen, AR Chandrasekaran. Biochemistry 60, 250-253 (2021). [PDF]
Nanoscale devices that can respond to external stimuli have potential applications in drug delivery, biosensing, and molecular computation. Construction using DNA has provided many such devices that can respond to cues such as nucleic acids, proteins, pH, light, or temperature. However, simultaneous control of molecular devices is still limited. Here, we present orthogonal control of DNA nanoswitches using physical (light) and biochemical (enzyme and nucleic acid) triggers. Each one of these triggers controls the reconfiguration of specific nanoswitches from locked to open states within a mixture and can be used in parallel to control a combination of nanoswitches. Such dynamic control over nanoscale devices allows the incorporation of tunable portions within larger structures as well as spatiotemporal control of DNA nanostructures.
DNA nanoswitch barcodes for multiplexed biomarker profiling
AR Chandrasekaran, M MacIsaac, J Vilcapoma, CH Hansen, D Yang, WP Wong, K Halvorsen. Nano Lett. 21, 469-475 (2021). [PDF]
Molecular biomarkers play a key role in the clinic, aiding in diagnostics and prognostics, and in the research laboratory, contributing to our basic understanding of diseases. Detecting multiple and diverse molecular biomarkers within a single accessible assay would have great utility, providing a more comprehensive picture for clinical evaluation and research, but is a challenge with standard methods. Here, we report programmable DNA nanoswitches for multiplexed detection of up to 6 biomarkers at once with each combination of biomarkers producing a unique barcode signature among 64 possibilities. As a defining feature of our method, we show “mixed multiplexing” for simultaneous barcoded detection of different types of biomolecules, for example, DNA, RNA, antibody, and protein in a single assay. To demonstrate clinical potential, we show multiplexed detection of a prostate cancer biomarker panel in serum that includes two microRNA sequences and prostate specific antigen.
Rapid one-step detection of SARS-CoV-2 RNA [News & Views]
AR Chandrasekaran, L Zhou, K Halvorsen. Nat. Biomed. Eng. 4, 1123-1124 (2020). [PDF]
A one-pot fluorescence-based assay detects SARS-CoV-2 RNA in under an hour with high sensitivity and sequence specificity.
A Wi-Fi live streaming Centrifuge Force Microscope for benchtop single-molecule experiments
JA Punnoose, A Hayden, L Zhou, K Halvorsen. Biophysical J. 119, 2231-2239 (2020). [PDF]
The ability to apply controlled forces to individual molecules has been revolutionary in shaping our understanding of biophysics in areas as diverse as dynamic bond strength, biological motor operation, and DNA replication. However, the methodology to perform single-molecule experiments has been and remains relatively inaccessible due to cost and complexity. In 2010, we introduced the Centrifuge Force Microscope (CFM) as a new platform for accessible and high-throughput single-molecule experimentation. The CFM consists of a rotating microscope where prescribed centrifugal forces can be applied to microsphere-tethered biomolecules. In this work, we develop and demonstrate a next-generation Wi-Fi CFM that offers unprecedented ease of use and flexibility in design. The modular CFM unit fits within a standard benchtop centrifuge and connects by Wi-Fi to a external computer for live control and streaming at near gigabit speeds. The use of commercial wireless hardware allows for flexibility in programming and provides a streamlined upgrade path as Wi-Fi technology improves. To facilitate ease of use, detailed build and setup instructions are provided, as well as LabVIEW based control software and MATLAB based analysis software. We demonstrate the analysis of force-dependent dissociation of short DNA duplexes of 7, 8, and 9 bp using the instrument. We showcase the sensitivity of the approach by resolving distinct dissociation kinetic rates for a 7 bp duplex where one G-C base pair is mutated to an A-T base pair.
Hybrid DNA/RNA nanostructures with 2′-5′ linkages
AR Chandrasekaran, J Mathivanan, P Ebrahimi, J Vilcapoma, AA Chen, K Halvorsen, J Sheng. Nanoscale 12, 21583-21590 (2020). [PDF]
Nucleic acid nanostructures with different chemical compositions have shown utility in biological applications as they provide additional assembly parameters and enhanced stability. The naturally occurring 2′-5′ linkage in RNA is thought to be a prebiotic analogue and has potential use in antisense therapeutics. Here, we report the first instance of DNA/RNA motifs containing 2′-5′ linkages. We synthesized and incorporated RNA strands with 2′-5′ linkages into different DNA motifs with varying number of branch points (a duplex, four arm junction, double crossover motif and tensegrity triangle motif). Using experimental characterization and molecular dynamics simulations, we show that hybrid DNA/RNA nanostructures can accommodate interspersed 2′-5′ linkages with relatively minor effect on the formation of these structures. Further, the modified nanostructures showed improved resistance to ribonuclease cleavage, indicating their potential use in the construction of robust drug delivery vehicles with prolonged stability in physiological conditions.
Programmable low-cost DNA-based platform for viral RNA detection.
L Zhou, AR Chandrasekaran, JA Punnoose, G Bonenfant, S Charles, O Levchenko, P Badu, C Cavaliere, CT Pager, K Halvorsen. Sci. Adv. 6, eabc6246 (2020). [PDF]
Detection of viruses is critical for controlling disease spread. Recent emerging viral threats including Zika virus, Ebola virus, and SARS-Cov-2 (responsible for COVID-19) highlight the cost and difficulty in responding rapidly. To address these challenges, we develop a platform for low-cost and rapid detection of viral RNA with DNA nanoswitches that mechanically reconfigure in response to specific viruses. Using Zika virus as a model system, we show non-enzymatic detection of viral RNA, with selective and multiplexed detection between related viruses and viral strains. For clinical-level sensitivity in biological fluids, we paired the assay with sample preparation using either RNA extraction or isothermal pre-amplification. Our assay requires minimal lab infrastructure, and is adaptable to other viruses, as demonstrated by quickly developing DNA nanoswitches to detect SARS-CoV-2 RNA in saliva. We expect further development and field implementation will improve our ability to detect emergent viral threats and ultimately limit their impact.
Nuclease degradation analysis of DNA nanostructures using gel electrophoresis
AR Chandrasekaran, K Halvorsen. Curr. Protoc. Nucleic Acid Chem. 82, e115 (2020). [PDF]
Custom‐built DNA nanostructures are now used in applications such as biosensing, molecular computation, biomolecular analysis, and drug delivery. While the functionality and biocompatibility of DNA makes DNA nanostructures useful in such applications, the field faces a challenge in making biostable DNA nanostructures. Being a natural material, DNA is most suited for biological applications, but is also easily degraded by nucleases. Several methods have been employed to study the nuclease degradation rates and enhancement of nuclease resistance. This protocol describes the use of gel electrophoresis to analyze the extent of nuclease degradation of DNA nanostructures and to report degradation times, kinetics of nuclease digestion, and evaluation of biostability enhancement factors.
Ribonuclease-Responsive DNA Nanoswitches
AR Chandrasekaran, R Trivedi, K Halvorsen. Cell Reports Phy. Sci. 1, 100117 (2020). [PDF]
DNA has been used in the construction of dynamic DNA devices that can reconfigure in the presence of external stimuli. These nanodevices have found uses in fields ranging from biomedical to materials science applications. Here, we report a DNA nanoswitch that can be reconfigured using ribonucleases (RNases) and explore two applications: biosensing and molecular computing. For biosensing, we show the detection of RNase H and other RNases in relevant biological fluids and temperatures, as well as inhibition by the known enzyme inhibitor kanamycin. For molecular computing, we show that RNases can be used to enable erasing, write protection, and erase-rewrite functionality for information-encoding DNA nanoswitches. The simplistic mix-and-read nature of the ribonuclease-activated DNA nanoswitches could facilitate their use in assays for identifying RNase contamination in biological samples or for the screening and characterization of RNase inhibitors.
Parallel poly(A) homo- and hetero-duplex formation detection with an adapted DNA nanoswitch technique.
MAG Pickard, KB Brylow, LA Cisco, MR Anecelle, ML Pershun, AR Chandrasekaran, K Halvorsen, ML Gleghorn. RNA 26: 1118-1130 (2020). [PDF]
Polyriboadenylic (poly(rA)) strands of sufficient length form parallel double helices in acidic and/or ammonium-containing conditions. Poly(rA) duplexes in acidic conditions are held together by A–A base-pairing also involving base interactions with the phosphate backbone. Traditional UV-melting studies of parallel poly(A) duplexes have typically examined homo-duplex formation of a single nucleic acid species in solution. We have adapted a technique utilizing a DNA nanoswitch that detects interaction of two different strands either with similar or differing lengths or modifications. Our method detected parallel duplex formation as a function of length, chemical modifications, and pH, and at a sensitivity that required over 100 fold less concentration of sample than prior UV-melting methods. While parallel polyriboadenylic acid and poly-2′-O-methyl-adenylic acid homo-duplexes formed, we did not detect homo-duplexes of polydeoxyriboadenylic acid strands or poly-locked nucleic acid (LNA)-adenylic strands. Importantly however, a poly-locked nucleic acid (LNA)-adenylic strand, as well as a poly-2′-O-methyl-adenylic strand formed a hetero-duplex with a polyriboadenylic strand. Overall, our work validates a new tool for studying parallel duplexes and reveals fundamental properties of poly(A) parallel duplex formation. Parallel duplexes may find use in DNA nanotechnology and in molecular biology applications such as a potential poly(rA) tail capture tool as an alternative to traditional oligo(dT) based purification.
Exceptional nuclease resistance of paranemic crossover (PX) DNA and crossover-dependent biostability of DNA motifs.
AR Chandrasekaran, J Vilcapoma, P Dey, SW Wong-Deyrup, BK Dey, K Halvorsen. J. Am. Chem. Soc. 142, 6814-6821 (2020). [PDF]
Inherent nanometer-sized features and molecular recognition properties make DNA a useful material in constructing nanoscale objects, with alluring applications in biosensing and drug delivery. However, DNA can be easily degraded by nucleases present in biological fluids, posing a considerable roadblock to realizing the full potential of DNA nanotechnology for biomedical applications. Here we investigated the nuclease resistance and biostability of the multi-stranded motif called paranemic crossover (PX) DNA and discovered a remarkable and previously unreported resistance to nucleases. We show that PX DNA has more than an order of magnitude increased resistance to degradation by DNase I, serum, and urine compared to double stranded DNA. We further demonstrate that the degradation resistance decreases monotonically as DNA crossovers are removed from the structure, suggesting that frequent DNA crossovers disrupt either the binding or catalysis of nucleases or both. Further, we show using mouse and human cell lines that PX DNA does not affect cell proliferation or interfere with biological processes such as myogenesis. These results have important implications for building DNA nanostructures with enhanced biostability, either by adopting PX-based architectures or by carefully engineering crossovers. We contend that such crossover-dependent nuclease resistance could potentially be used to add “tunable biostability” to the many features of DNA nanotechnology.
DNA nanotechnology in the undergraduate laboratory: Analysis of molecular topology using DNA nanoswitches
JA Punnoose, K Halvorsen, AR Chandrasekaran. J. Chem. Edu. 97, 1448-1453 (2020). [PDF]
There is a disconnect between the cutting-edge research done in academic labs, such as nanotechnology, and what is taught in undergraduate labs. In the current undergraduate curriculum, very few students get a chance to do hands-on experiments in nanotechnology-related experiments most of which are through selective undergraduate research programs. In most cases, complicated synthesis procedures, expensive reagents, and requirement of specific instrumentation prevent broad adaptation of nanotechnology-based experiments to laboratory courses. DNA, being a nanoscale molecule, has recently been used in bottom-up nanotechnology with applications in sensing, nano-robotics, and computing. In this article, we propose a simple experiment involving the synthesis of a DNA nanoswitch that can change its shape from a linear “off” state to a looped “on” state in the presence of a target DNA molecule. The experiment also demonstrates the programmable topology of the looped state of the nanoswitch and its effect on gel migration. The experiment is easy to adapt in an undergraduate laboratory, requires only agarose gel electrophoresis, a minimal set-up cost for materials, and can be completed in a 3-hour time frame.
How to perform miRacles: A step‐by‐step microRNA detection protocol using DNA nanoswitches.
AR Chandrasekaran, BK Dey, K Halvorsen. Curr. Protoc. Mol. Biol. 130, e114 (2020). [PDF]
Viral detection is critical for controlling disease spread and progression. Recent emerging threats including the Zika and Ebola virus outbreaks highlight the cost and difficulty in responding rapidly. In low-resource areas, a key obstacle is quick and accurate detection of viruses near the point of care. To address these challenges, we develop a platform for low-cost and rapid detection of viral RNA with DNA nanoswitches designed to mechanically reconfigure in response to specific viruses. Using Zika virus as a model system, we show non-enzymatic detection of viral RNA to the attomole level, with selective and multiplexed detection between related viruses and viral strains. For clinical-level sensitivity in biological fluids, we paired the assay with a sample preparation step using either RNA extraction or isothermal pre-amplification. Our assay can be performed with minimal or no lab infrastructure, and is readily adaptable (with ~24-hour development time) to detect other viruses. Given this versatility, we expect that further development and field implementation will improve our ability to detect emergent viral threats and ultimately limit their impact.
Click and photo-release dual-functional nucleic acid nanostructures.
VA Valsangkar, AR Chandrasekaran, L Zhou, S Mao, GW Lee, M Kizer, X Wang, K Halvorsen, J Sheng. Chem. Commun. 55, 9709-9712 (2019). [PDF]
We functionalize nucleic acid nanostructures with click chemistry (for attachment of cargos) and a photocleavable linker (for release). We demonstrate cargo attachment using a fluorescein dye and release using UV trigger from an RNA three-way junction, a DNA star motif and a DNA tetrahedron. Such multifunctional nucleic acid nanostructures have potential in targeted drug delivery.
DNA nanotechnology approaches for microRNA detection and diagnosis.
AR Chandrasekaran, JA Punnoose, L Zhou, P Dey, BK Dey, K Halvorsen. Nucleic Acids Res. 47, 10489-10505 (2019). [PDF]
MicroRNAs are involved in the crucial processes of development and diseases and have emerged as a new class of biomarkers. The field of DNA nanotechnology has shown great promise in the creation of novel microRNA biosensors that have utility in lab-based biosensing and potential for disease diagnostics. In this Survey and Summary, we explore and review DNA nanotechnology approaches for microRNA detection, surveying the literature for microRNA detection in three main areas of DNA nanostructures: DNA tetrahedra, DNA origami, and DNA devices and motifs. We take a critical look at the reviewed approaches, advantages and disadvantages of these methods in general, and a critical comparison of specific approaches. We conclude with a brief outlook on the future of DNA nanotechnology in biosensing for microRNA and beyond.
Integration of a photocleavable element into DNA nanoswitches.
AR Chandrasekaran, JA Punnoose, V Valsangkar, J Sheng, K Halvorsen. Chem. Commun. 55, 6587-6590 (2019). [PDF]
Reconfigurable DNA nanostructures can be designed to respond to external stimuli such as nucleic acids, pH, small molecules and enzymes. In this study, we incorporated photocleavable linkers in DNA strands that trigger a conformational change in binary DNA nanoswitches. We demonstrate control of the output using UV light, with potential applications in biosensing and molecular computation.
Cellular microRNA detection with miRacles: microRNA- activated conditional looping of engineered switches.
AR Chandrasekaran, M MacIsaac, P Dey, O Levchenko, L Zhou, M Andres, BK Dey, K Halvorsen. Science Adv. 5, eaau9443 334631 (2019). [PDF]
MicroRNAs are short non-coding regulatory RNAs that are increasingly used as disease biomarkers. Detection of microRNAs can be arduous and expensive, and often requires amplification, labeling, or radioactive probes. Here we report a single-step, non- enzymatic detection assay using conformationally responsive DNA nanoswitches. Termed miRacles (microRNA activated conditional looping of engineered switches), our assay has sub-attomole sensitivity and single-nucleotide specificity using an agarose gel electrophoresis readout. We detect cellular microRNAs from nanogram-scale RNA extracts of differentiating muscle cells, and demonstrate multiplexed detection of several microRNAs from one biological sample. We demonstrate one-hour detection without expensive equipment or reagents, making this assay a compelling alternative to qPCR and Northern blotting.
Controlled disassembly of a DNA tetrahedron using strand displacement.
AR Chandrasekaran, K Halvorsen. Nanoscale Adv. 1, 969-972 (2018). [PDF]
In this study, we assembled a DNA tetrahedron containing single stranded extensions in the middle of the struts. Using these extensions as toeholds, the tetrahedron can be disassembled by nucleic acid triggers via strand displacement. The release mechanism is sequence specific, is functional in biological fluids such as serum and urine, and the kinetics of the disassembly process can be controlled by different molar ratios of the release strand. Such DNA nanostructures that respond to external stimuli have potential use in biosensing and drug delivery, and we demonstrate proof-of-concept of this approach for microRNA detection.
A ‘smart’tube holder enables real-time sample monitoring in a standard lab centrifuge.
T Hoang, N Moskwa, K Halvorsen. PLoS One 13, e0195907 (2018). [PDF]
The centrifuge is among the oldest and most widely used pieces of laboratory equipment, with significant applications that include clinical diagnostics and biomedical research. A major limitation of laboratory centrifuges is their “black box” nature, limiting sample observation to before and after centrifugation. Thus, optimized protocols require significant trial and error, while unoptimized protocols waste time by centrifuging longer than necessary or material due to incomplete sedimentation. Here, we developed an instrumented centrifuge tube receptacle compatible with several commercial benchtop centrifuges that can provide real-time sample analysis during centrifugation. We demonstrated the system by monitoring cell separations during centrifugation for different spin speeds, concentrations, buffers, cell types, and temperatures. We verified an adaptation where complete sedimentation turned off the centrifuge and notified the user by a text message. Our system adds new functionality to existing laboratory centrifuges, saving users time and providing useful feedback.
Addressable configurations of DNA nanostructures for rewritable memory.
AR Chandrasekaran,* O Levchenko, D Patel, M MacIsaac and K Halvorsen. Nucleic Acids Res. 45, 11459-11465 (2017) [PDF].
DNA serves as nature's information storage molecule, and has been the primary focus of engineered systems for biological computing and data storage. Here we combine recent efforts in DNA self-assembly and toehold-mediated strand displacement to develop a rewritable multi-bit DNA memory system. The system operates by encoding information in distinct and reversible conformations of a DNA nanoswitch and decoding by gel electrophoresis. We demonstrate a 5-bit system capable of writing, erasing, and rewriting binary representations of alphanumeric symbols, as well as compatibility with ‘OR’ and ‘AND’ logic operations. Our strategy is simple to implement, requiring only a single mixing step at room temperature for each operation and standard gel electrophoresis to read the data. We envision such systems could find use in covert product labeling and barcoding, as well as secure messaging and authentication when combined with previously developed encryption strategies. Ultimately, this type of memory has exciting potential in biomedical sciences as data storage can be coupled to sensing of biological molecules.
Shear dependent LC purification of an engineered DNA nanoswitch and implications for DNA origami.
K Halvorsen, M Kizer, X Wang, AR Chandrasekaran and M Basanta-Sanchez. Anal. Chem. 89, 5673-5677 (2017) [PDF].
As DNA nanotechnology matures, there is increasing need for fast, reliable, and automated purification methods. Here, we develop UHPLC methods to purify self-assembled DNA nanoswitches, which are formed using DNA origami approaches and are designed to change conformations in response to a binding partner. We found that shear degradation hindered LC purification of the DNA nanoswitches, removing oligonucleotides from the scaffold strand and causing loss of function. However, proper choice of column, flow rate, and buffers enabled robust and automated purification of DNA nanoswitches without loss of function in under a half hour. Applying our approach to DNA origami structures, we found that ∼400 nm long nanotubes degraded under the gentlest flow conditions while ∼40 nm diameter nanospheres remained intact even under aggressive conditions. These examples show how fluid stresses can affect different DNA nanostructures during LC purification and suggest that shear forces may be relevant for some applications of DNA nanotechnology. Further development of this approach could lead to fast and automated purification of DNA nanostructures of various shapes and sizes, which would be an important advance for the field.
Click-based functionalization of a 2'-O-propargyl-modified branched DNA nanostructure.
V Valsangkar, AR Chandrasekaran, R Wang, P Haruehanroengra, O Levchenko, K Halvorsen and J Sheng. J. Mater. Chem. B 5, 2074-2077 (2017) [PDF].
DNA has emerged as a versatile building block for programmable self-assembly. DNA-based nanostructures have been widely applied in biosensing, bioimaging, drug delivery, molecular computation and macromolecular scaffolding. A variety of strategies have been developed to functionalize these nanostructures. In this study, we report a facile click-based strategy to incorporate a metal chelating ligand and a fluorescent tag into a three-point-star DNA tile containing 2′-O-propargyl groups. Such a strategy opens up the possibility of functionalizing pre-assembled DNA strands to construct platforms for metal or drug delivery.
A wireless centrifuge force microscope (CFM) enables multiplexed single-molecule experiments in a commercial centrifuge.
T Hoang, DS Patel, K Halvorsen. Rev. Sci. Instrum. 87, 083705 (2016) [PDF].
The centrifuge force microscope (CFM) was recently introduced as a platform for massively parallel single-molecule manipulation and analysis. Here we developed a low-cost and self-contained CFM module that works directly within a commercial centrifuge, greatly improving accessibility and ease of use. Our instrument incorporates research grade video microscopy, a power source, a computer, and wireless transmission capability to simultaneously monitor many individually tethered microspheres. We validated the instrument by performing single-molecule force shearing of short DNA duplexes. For a 7 bp duplex, we observed over 1000 dissociation events due to force dependent shearing from 2 pN to 12 pN with dissociation times in the range of 10-100 s. We extended the measurement to a 10 bp duplex, applying a 12 pN force clamp and directly observing single-molecule dissociation over an 85 min experiment. Our new CFM module facilitates simple and inexpensive experiments that dramatically improve access to single-molecule analysis.
Beyond the fold: Emerging biological applications of DNA origami.
AR Chandrasekaran, N Anderson, M Kizer, K Halvorsen and X Wang, ChemBioChem 17, 1081-1089 (2016) [PDF].
The use of DNA as a material for nanoscale construction has blossomed in the past decade. This is largely attributable to the DNA origami technique, which has enabled construction of nanostructures ranging from simple two‐dimensional sheets to complex three‐dimensional objects with defined curves and edges. These structures are amenable to site‐specific functionalization with nanometer precision, and have been shown to exhibit cellular biocompatibility and permeability. The DNA origami technique has already found widespread use in a variety of emerging biological applications such as biosensing, enzyme cascades, biomolecular analysis, biomimetics, and drug delivery. We highlight a few of these applications and comments on the prospects for this rapidly expanding field of research.
Complex thermodynamic behavior of single-stranded nucleic acid adsorption to graphene surfaces.
SV Ranganathan, K Halvorsen, CA Myers, NM Robertson, MV Yigit, AA Chen. Langmuir 32, 6028-6034 (2016) [PDF].
Graphene oxide has shown promise as a biosensor due to its preferential absorption of single-stranded polynucleotides and fluorescence quenching properties. The rational design of these biosensors, however, requires an improved understanding of the binding thermodynamics and ultimately a predictive model of sequence-specific binding. Toward these goals, here we directly measured the binding of nucleosides and oligonucleotides to graphene oxide nanoparticles using isothermal titration calorimetry and used the results to develop molecular models of graphene–nucleic acid interactions. Experimental and computational results from this study set the platform for informed design of graphene-based biosensors, further increasing their potential and application.
Evolution of DNA origami scaffolds.
AR Chandrasekaran, M Pushpanathan, K Halvorsen. Mater. Lett. 170, 221-224 (2016) [PDF].
Nanoscale materials made using DNA have been increasingly used for applications ranging from biosensors to nanoelectronics. Specifically, DNA origami – where one long single-stranded DNA scaffold is folded into nanoscale shapes and structures using short ‘staple’ oligonucleotides – typically relies on a single-stranded DNA scaffold derived from a viral genome. The sizes of structures that are made rely on the length of the scaffold strand; the most frequently used DNA scaffold is the single-stranded 7249-nucleotide circular M13mp18 genome. Modern techniques used in genome tailoring are now widely exploited for the creation of DNA scaffolds of various lengths for use in DNA origami. DNA scaffolds of lengths ranging from ~700-nucleotides to ~51,000 nucleotides have been prepared using biotechniques such as polymerase chain reaction, a combination of site-directed mutagenesis and site- and ligation independent cloning, and using the molecular toolbox of restriction and ligation enzymes. Such tailor-made DNA sca ffolds allow the creation of origami nanostructures of desired sizes.
Multiplexed single-molecule force spectroscopy using a centrifuge.
D Yang, A Ward, K Halvorsen, WP Wong. Nat. Commun. 7, 11026 (2016) [PDF].
We present a miniature centrifuge force microscope (CFM) that repurposes a benchtop centrifuge for high-throughput single-molecule experiments with high-resolution particle tracking, a large force range, temperature control and simple push-button operation. Incorporating DNA nanoswitches to enable repeated interrogation by force of single molecular pairs, we demonstrate increased throughput, reliability and the ability to characterize population heterogeneity. We perform spatiotemporally multiplexed experiments to collect 1,863 bond rupture statistics from 538 traceable molecular pairs in a single experiment, and show that 2 populations of DNA zippers can be distinguished using per-molecule statistics to reduce noise.
Programmable DNA nanoswitches for detection of nucleic acid sequences.
AR Chandrasekaran, J Zavala and K Halvorsen, ACS Sens. 1, 120-123 (2016) [PDF].
Detection of nucleic acid sequences is important for applications such as medicine and forensics, but many detection strategies involve multiple time-consuming steps or require expensive lab equipment. Here we report a programmable DNA nanoswitch that undergoes a predefined conformational change upon binding a target sequence, flipping the switch from a linear “off” state to a looped “on” state. The presence of the target sequence is determined without amplification using standard gel electrophoresis to separate the on and off states. We showed sensitivity into the low picomolar range, as well as detection of a single target sequence from both a randomized pool of high concentration oligonucleotides and from a solution of fetal bovine serum (FBS), with no false positive detection in either case. By leveraging the already ubiquitous technique of gel electrophoresis, our low cost approach will be especially accessible to researchers in the biomedical sciences.
DNA nanoswitches: a quantitative platform for gel-based biomolecular interaction analysis.
MA Koussa, K Halvorsen, A Ward, WP Wong. Nat. Meth. 12, 123-126 (2015) [PDF].
We introduce a nanoscale experimental platform that enables kinetic and equilibrium measurements of a wide range of molecular interactions using a gel electrophoresis readout. Programmable, self-assembled DNA nanoswitches serve both as templates for positioning molecules and as sensitive, quantitative reporters of molecular association and dissociation. We demonstrated this low-cost, versatile, 'lab-on-a-molecule' system by characterizing ten different interactions, including a complex four-body interaction with five discernible states.
Cross-platform comparison of nucleic acid hybridization: Toward quantitative reference standards.
K Halvorsen, PF Agris. Anal. Biochem. 465, 127-133 (2014) [PDF].
Measuring interactions between biological molecules is vitally important to both basic and applied research as well as development of pharmaceuticals. Although a wide and growing range of techniques is available to measure various kinetic and thermodynamic properties of interacting biomolecules, it can be difficult to compare data across techniques of different laboratories and personnel or even across different instruments using the same technique. Here we evaluate relevant biological interactions based on complementary DNA and RNA oligonucleotides that could be used as reference standards for many experimental systems. We measured thermodynamics of duplex formation using isothermal titration calorimetry, differential scanning calorimetry, and ultraviolet-visible (UV-vis) monitored denaturation/renaturation. These standards can be used to validate results, compare data from disparate techniques, act as a teaching tool for laboratory classes, or potentially to calibrate instruments. The RNA and DNA standards have many attractive features, including low cost, high purity, easily measurable concentrations, and minimal handling concerns, making them ideal for use as a reference material.
Bioinspired multivalent DNA network for capture and release of cells.
W Zhao, CH Cui, S Bose, D Guo, C Shen, WP Wong, K Halvorsen, OC Farokhzad, GSL Teo, JA Phillips, DM Dorfman, R Karnik, JM Karp. PNAS 109, 19626-19631 (2012) [PDF].
Capture and isolation of flowing cells and particulates from body fluids has enormous implications in diagnosis, monitoring, and drug testing, yet monovalent adhesion molecules used for this purpose result in inefficient cell capture and difficulty in retrieving the captured cells. Inspired by marine creatures that present long tentacles containing multiple adhesive domains to effectively capture flowing food particulates, we developed a platform approach to capture and isolate cells using a 3D DNA network comprising repeating adhesive aptamer domains that extend over tens of micrometers into the solution. The DNA network was synthesized from a microfluidic surface by rolling circle amplification where critical parameters, including DNA graft density, length, and sequence, could readily be tailored. Using an aptamer that binds to protein tyrosine kinase-7 (PTK7) that is overexpressed on many human cancer cells, we demonstrate that the 3D DNA network significantly enhances the capture efficiency of lymphoblast CCRF-CEM cells over monovalent aptamers and antibodies, yet maintains a high purity of the captured cells. When incorporated in a herringbone microfluidic device, the 3D DNA network not only possessed significantly higher capture efficiency than monovalent aptamers and antibodies, but also outperformed previously reported cell-capture microfluidic devices at high flow rates. This work suggests that 3D DNA networks may have broad implications for detection and isolation of cells and other bioparticles.
Binary DNA nanostructures for data encryption.
K Halvorsen, WP Wong. PloS One 7, e44212 (2012) [PDF].
We present a simple and secure system for encrypting and decrypting information using DNA self-assembly. Binary data is encoded in the geometry of DNA nanostructures with two distinct conformations. Removing or leaving out a single component reduces these structures to an encrypted solution of ssDNA, whereas adding back this missing “decryption key” causes the spontaneous formation of the message through self-assembly, enabling rapid read out via gel electrophoresis. Applications include authentication, secure messaging, and barcoding.
Physical manipulation of the Escherichia coli chromosome reveals its soft nature.
J Pelletier, K Halvorsen, BY Ha, R Paparcone, SJ Sandler, CL Woldringh, WP Wong, S Jun. PNAS 109, E2649-E2656 (2012) [PDF].
Replicating bacterial chromosomes continuously demix from each other and segregate within a compact volume inside the cell called the nucleoid. Although many proteins involved in this process have been identified, the nature of the global forces that shape and segregate the chromosomes has remained unclear because of limited knowledge of the micromechanical properties of the chromosome. In this work, we demonstrate experimentally the fundamentally soft nature of the bacterial chromosome and the entropic forces that can compact it in a crowded intracellular environment. We developed a unique “micropiston” and measured the force-compression behavior of single Escherichia coli chromosomes in confinement. Our data show that forces on the order of 100 pN and free energies on the order of 105 kBT are sufficient to compress the chromosome to its in vivo size. For comparison, the pressure required to hold the chromosome at this size is a thousand-fold smaller than the surrounding turgor pressure inside the cell. Furthermore, by manipulation of molecular crowding conditions (entropic forces), we were able to observe in real time fast (approximately 10 s), abrupt, reversible, and repeatable compaction–decompaction cycles of individual chromosomes in confinement. In contrast, we observed much slower dissociation kinetics of a histone-like protein HU from the whole chromosome during its in vivo to in vitro transition. These results for the first time provide quantitative, experimental support for a physical model in which the bacterial chromosome behaves as a loaded entropic spring in vivo.
Nanoengineering a single-molecule mechanical switch using DNA self-assembly.
K Halvorsen, D Schaak, WP Wong. Nanotechnology 22, 494005 (2011) [PDF].
The ability to manipulate and observe single biological molecules has led to both fundamental scientific discoveries and new methods in nanoscale engineering. A common challenge in many single-molecule experiments is reliably linking molecules to surfaces, and identifying their interactions. We have met this challenge by nanoengineering a novel DNA-based linker that behaves as a force-activated switch, providing a molecular signature that can eliminate errant data arising from non-specific and multiple interactions. By integrating a receptor and ligand into a single piece of DNA using DNA self-assembly, a single tether can be positively identified by force–extension behavior, and receptor–ligand unbinding easily identified by a sudden increase in tether length. Additionally, under proper conditions the exact same pair of molecules can be repeatedly bound and unbound. Our approach is simple, versatile and modular, and can be easily implemented using standard commercial reagents and laboratory equipment. In addition to improving the reliability and accuracy of force measurements, this single-molecule mechanical switch paves the way for high-throughput serial measurements, single-molecule on-rate studies, and investigations of population heterogeneity.
Massively parallel single-molecule manipulation using centrifugal force.
K Halvorsen, WP Wong. Biophy J. 98, L53-L55 (2010) [PDF].
Precise manipulation of single molecules has already led to remarkable insights in physics, chemistry, biology, and medicine. However, widespread adoption of single-molecule techniques has been impeded by equipment cost and the laborious nature of making measurements one molecule at a time. We have solved these issues by developing an approach that enables massively parallel single-molecule force measurements using centrifugal force. This approach is realized in an instrument that we call the centrifuge force microscope in which objects in an orbiting sample are subjected to a calibration-free, macroscopically uniform force-field while their micro-to-nanoscopic motions are observed. We demonstrate high-throughput single-molecule force spectroscopy with this technique by performing thousands of rupture experiments in parallel, characterizing force-dependent unbinding kinetics of an antibody-antigen pair in minutes rather than days. Additionally, we verify the force accuracy of the instrument by measuring the well-established DNA overstretching transition at 66 ± 3 pN. With significant benefits in efficiency, cost, simplicity, and versatility, single-molecule centrifugation has the potential to expand single-molecule experimentation to a wider range of researchers and experimental systems.
High-precision microsphere sorting using velocity sedimentation.
D Cheng, K Halvorsen, WP Wong. Rev. Sci. Instrum. 81, 026106 (2010) [PDF].
Monodisperse populations of microspheres are desirable for a variety of research and industrial applications, but many desirable sizes and materials can be difficult to synthesize and have limited commercial availability. In this paper, we present an effective, straightforward, and low cost method for sorting polydisperse microspheres into many separate monodisperse samples. The basic approach is to use velocity sedimentation through a density gradient in a long vertical column, followed by carefully targeted extraction. We demonstrate this technique by reducing the coefficient of variation of melamine microspheres from 13% to 1%–4% and glass microspheres from 35% to 3%–8%. This simple and inexpensive method can be used to sort microspheres of many sizes and materials, and is easily scalable, opening the possibility of cheap, monodisperse microspheres.
Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor.
X Zhang, K Halvorsen, CZ Zhang, WP Wong, TA Springer. Science 324, 1330-1334 (2009) [PDF].
Von Willebrand factor (VWF) is secreted as ultralarge multimers that are cleaved in the A2 domain by the metalloprotease ADAMTS13 to give smaller multimers. Cleaved VWF is activated by hydrodynamic forces found in arteriolar bleeding to promote hemostasis, whereas uncleaved VWF is activated at lower, physiologic shear stresses and causes thrombosis. Single-molecule experiments demonstrate that elongational forces in the range experienced by VWF in the vasculature unfold the A2 domain, and only the unfolded A2 domain is cleaved by ADAMTS13. In shear flow, tensile force on a VWF multimer increases with the square of multimer length and is highest at the middle, providing an efficient mechanism for homeostatic regulation of VWF size distribution by force-induced A2 unfolding and cleavage by ADAMTS13, as well as providing a counterbalance for VWF-mediated platelet aggregation.
Beyond the frame rate: Measuring high-frequency fluctuations with light-intensity modulation.
WP Wong, K Halvorsen. Opt. Lett. 34, 277-279 (2009) [PDF].
Power spectral density measurements of any sampled signal are typically restricted by both acquisition rate and frequency response limitations of instruments, which can be particularly prohibitive for video-based measurements. We have developed a new method called Intensity Modulation Spectral Analysis (IMSA) that circumvents these limitations, dramatically extending the effective detection bandwidth. We demonstrate this by video-tracking an optically-trapped microsphere while oscillating an LED illumination source. This approach allows us to quantify fluctuations of the microsphere at frequencies over 10 times higher than the Nyquist frequency, mimicking a significantly higher frame rate.
A new approach to analysis of single-molecule force measurements.
E Evans, K Halvorsen, K Kinoshita, WP Wong. Handbook of Single-Molecule Biophysics, 571-589 (2009).
A common aim in probing single molecular bonds or the structural stability of proteins is to measure the kinetic rates at which a bond dissociates or a protein changes conformation under conditions of changing force. Using sample data taken from tests of ligand–receptor unbinding and protein unfolding/refolding, we show that populations of “single molecule” events, arranged into statistical arrays expressing the numbers of bonds or initial conformers remaining as a function of time and cumulated into histograms of transitions over fixed time increments, provide the bases for a model-independent assay of the kinetic rates of transition throughout the course of an experiment. Most important, this assay for kinetic rates can be employed with any deterministic mode of force spectroscopy, whether the pulling force increases or decreases with time.
Imaging biomolecular interactions by fast three-dimensional tracking of laser-confined carrier particles.
V Heinrich, WP Wong, K Halvorsen, E Evans. Langmuir 24, 1194-1203 (2008) [PDF].
The quantitative study of the near-equilibrium structural behavior of individual biomolecules requires high-resolution experimental approaches with longtime stability. We present a new technique to explore the dynamics of weak intramolecular interactions. It is based on the analysis of the 3D Brownian fluctuations of a laser-confined glass bead that is tethered to a flat surface by the biomolecule of interest. A continuous autofocusing mechanism allows us to maintain or adjust the height of the optical trap with nanometer accuracy over long periods of time. The resulting remarkably stable trapping potential adds a well-defined femto-to-piconewton force bias to the energy landscape of molecular configurations. A combination of optical interferometry and advanced pattern-tracking algorithms provides the 3D bead positions with nanometer spatial and >120 Hz temporal resolution. The analysis of accumulated 3D positions has allowed us not only to identify small single biomolecules but also to characterize their nanomechanical behavior, for example, the force−extension relations of short oligonucleotides and the unfolding/refolding transitions of small protein tethers.
The effect of integration time on fluctuation measurements: calibrating an optical trap in the presence of motion blur.
WP Wong, K Halvorsen. Opt. Express 14, 12517-12531 (2006) [PDF].
Dynamical instrument limitations, such as finite detection bandwidth, do not simply add statistical errors to fluctuation measurements, but can create significant systematic biases that affect the measurement of steady-state properties. Such effects must be considered when calibrating ultra-sensitive force probes by analyzing the observed Brownian fluctuations. In this article, we present a novel method for extracting the true spring constant and diffusion coefficient of a harmonically confined Brownian particle that extends the standard equipartition and power spectrum techniques to account for video-image motion blur. These results are confirmed both numerically with a Brownian dynamics simulation, and experimentally with laser optical tweezers.